Wiring up the brain’s trillions of circuit connections is an enormous job performed by a huge crew of molecules. Among the less understood members are circular RNAs, transcripts from DNA that assume a closed loop shape. A new study by a team of neuroscientists centered at MIT shows that one such circular RNA from the Homer gene, “circHomer1,” takes on a significant and somewhat surprising role in how the developing brains of mice formed connections, or “synapses,” in the visual system.
In the study, the team demonstrates that interfering with circHomer1 not only undermined the normal development of synapses, but also delayed visual system neurons from making the expected adjustments when the scientists performed a classic experiment of temporarily blocking vision through one eye, a protocol called “monocular deprivation” (MD).
“It is used to build synapses, for sure,” said co-senior author Mriganka Sur, Newton Professor in The Picower Institute for Learning and Memory and MIT’s Department of Brain and Cognitive Sciences. “When you knock it down, the synapses, at least structurally, are not fully built. Then, after monocular deprivation, when the dendritic spines housing synapses normally should shrink, as should responses from the blocked eye, knocking down circHomer1 prevented that for three days.”
The lead authors of the study published in iScience are Sur Lab postdoc Kyle Jenks, former MIT graduate student Marvin Nayan, and graduate student Ying Cai in the lab of co-senior author and former Sur Lab postdoc Jacque Pak Kan Ip, at the Chinese University of Hong Kong.
Homing in on Homer
A linear RNA transcribed from the Homer gene (“Homer1a”), is rapidly and transiently upregulated in neurons in response to various forms of neuronal activation. Homer1a disrupts synaptic scaffolds that anchor glutamate receptors, weakening “excitatory” synapse function, Sur said. But over the last decade or so, Sur’s lab has also been curious about how other forms of RNA affect how synapses are built, particularly in response to nervous system activity (such as visual input) during development. The brain’s ability to adjust its networks to accommodate activity, an attribute called “plasticity,” is crucial to development, learning and memory.
The new study therefore began with an unbiased screen for RNAs, circular and linear, that showed significant differences in their degree of expression when the researchers performed the MD experiment. The screen showed that 73 circular RNAs were differently expressed. What made circHomer1 stand out in particular was that its expression increased for the first three days of MD even as Homer1a’s expression decreased during that same time. By a week of MD, however, circHomer1 expression had also decreased.
Intrigued, the team measured circHomer1’s expression levels during normal development and saw that it increases significantly at the start of the “critical period” when the brain undergoes a lot of remodeling to account for experience. Homer1a expression, by contrast, didn’t increase until near the end of the critical period. In general, Jenks said, the independent trajectories of the linear and circular forms of the RNA were notable.
Knocking out circHomer1
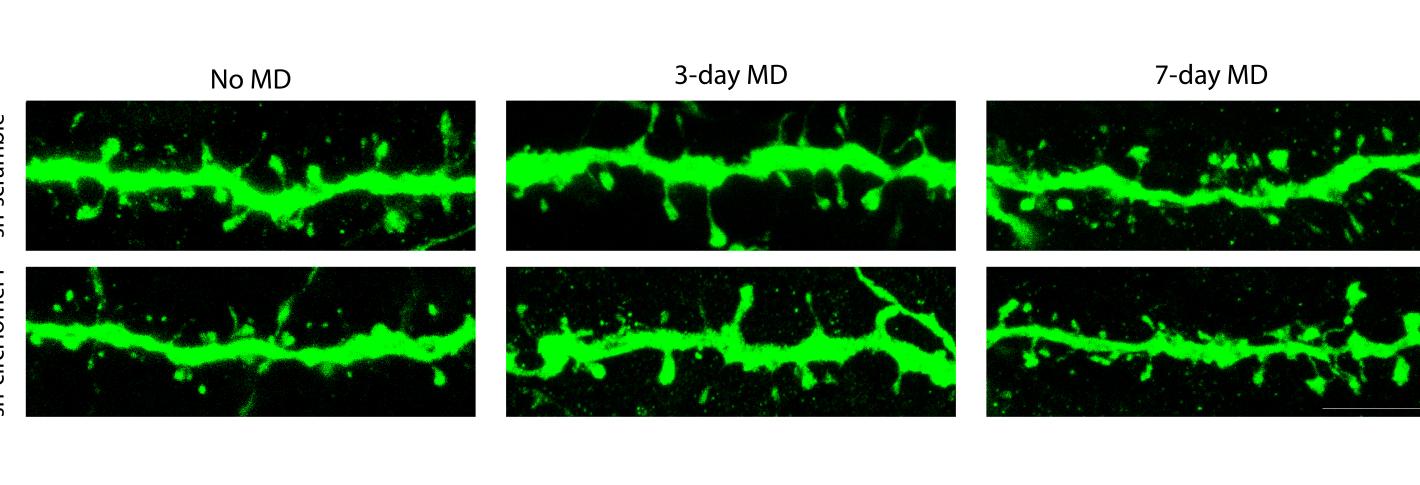
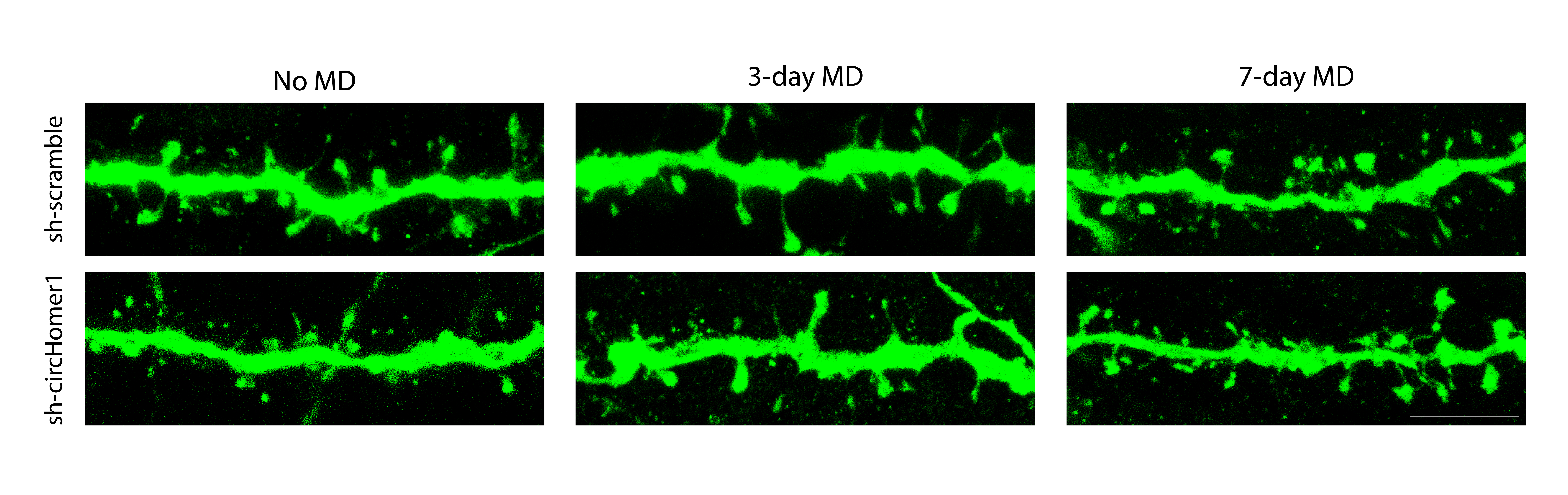
To see how circHomer1 might affect synapse development, they devised a way to interfere with its activity. That enabled them to make several observations about what happens to visual system neurons under four conditions: with or without circHomer1, and with or without MD.
In mice who didn’t undergo MD, they found that knocking out circHomer1 prevented the maturation of the spine structures on neural dendrites that host synapses. In contrast, spine maturation proceeded normally in non-MD mice in which circHomer1 functioned without interference.
Meanwhile, in mice that experienced MD with normal circHomer1 function, measures of neural responsiveness to visual input showed that the neurons serving the deprived eye showed the expected drop-off, but did not in mice where circHomer1 was disabled. When the team examined dendrites of neurons in mice with normal circHomer1, spines shrank (as expected), alongside the drop-off of visual input. But in MD mice with circHomer1 knocked out, this shrinkage didn’t happen for the first three days, suggesting that circHomer1 is necessary for the expected response to MD early on. Moreover, in the MD mice with circHomer1 knocked out, there was less removal of proteins called AMPA receptors, which normally facilitates spine shrinkage.
What surprised the team was that none of the effects of circHomer1 knockout were apparent by day 7 of MD. It’s as if another unmeasured mechanism kicked in to complete the brain’s response to MD when circHomer1 couldn’t accomplish the job.
The study therefore raises a number of new questions. For example, what mechanism compensates under MD conditions for the loss of circHomer1 by day 7? What does circHomer1 act on during normal conditions to affect synapse development? And how do linear Homer1’s and circHomer1 interact, if at all?
But the study does clearly demonstrate that circHomer1 plays a consequential role in synapse development. Its role is likely not limited to the visual cortex, where the current study focuses. Study co-author Nikolaos Mellios, a former Sur lab member and now founder of Circular Genomics Inc. in San Diego, has been studying circHomer1 in other brain regions, such as the prefrontal cortex.
In addition to Jenks, Cai, Nayan, Mellios, Sur and Ip, the paper’s other authors are Katya Tsimring, Keji Li, Jose Zepeda, Gregg Heller, Chloe Delepine, Jennifer Shih, Shiyang Yuan, Yao Zhu, Ye Wang, Yangyang Duan, Amy Fu, Taeyun Ku, Dae Hee Yun, and Kwanghun Chung.
This work was supported by NIH grants R01MH085802, R01EY028219, F31EY033649, F32EY032756, The Simons Foundation Autism Research Initiative through the Simons Center for the Social Brain, MIT, the National Research Foundation of Korea (RS-2023-00264980;, the Hong Kong Research Grants Council Early Career Scheme (24117220), General Research Fund (14117221), Area of Excellence Scheme (AoE/M-604/16), Theme-based Research Scheme (T13-605/18-W), Lo Kwee-Seong Biomedical Research Fund, Faculty Innovation Awards from the Faculty of Medicine CUHK (FIA2020/A/04), a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation, the Freedom Together Foundation and The Picower Institute for Learning and Memory.