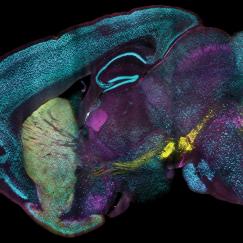
“It will be the first subcellular resolution, true volumetric human brain map,” said Chung, assistant professor in the Picower Institute, the Institute for Medical Engineering and Science and the Department of Chemical Engineering. “It will be like Google Maps. You will be able to drill all the way down to see what’s inside of individual ‘buildings’ and all the way up to see the ‘Earth’.”
Except in this simile the “buildings” are cells and the “Earth” is a whole human brain with all of its complex integration of circuits and regions. Truly a map – not just a detailed picture – the technology Chung’s team plans to implement will resolve specific proteins in individual cells, allowing scientists to identify their type and function. This is the equivalent of Google Maps not just showing you a picture of two MIT buildings across from each other on Vassar Street, but also revealing that in Building 46 there are neuroscientists and in the Stata Center there are roboticists.
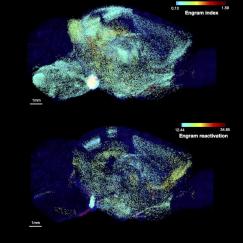
Producing a human brain map of this unprecedented scale, detail and multidimensionality could reveal entirely new types of brain cells and previously unknown connections, Chung said. Moreover, after the team implements all the technologies and methods needed to accomplish the first map, costs will invariably begin to come down, potentially leading to a revolution in brain mapping akin to genome sequencing.
“This will be the first demonstration, but after we have achieved this, we will be able to map many more brains with all sorts of neurological disorders and with decades-long medical records and functional data,” Chung said. “There are hundreds of brain banks worldwide and many tens of thousands of brains available. If we can map many brains, we will be able to learn a lot.”
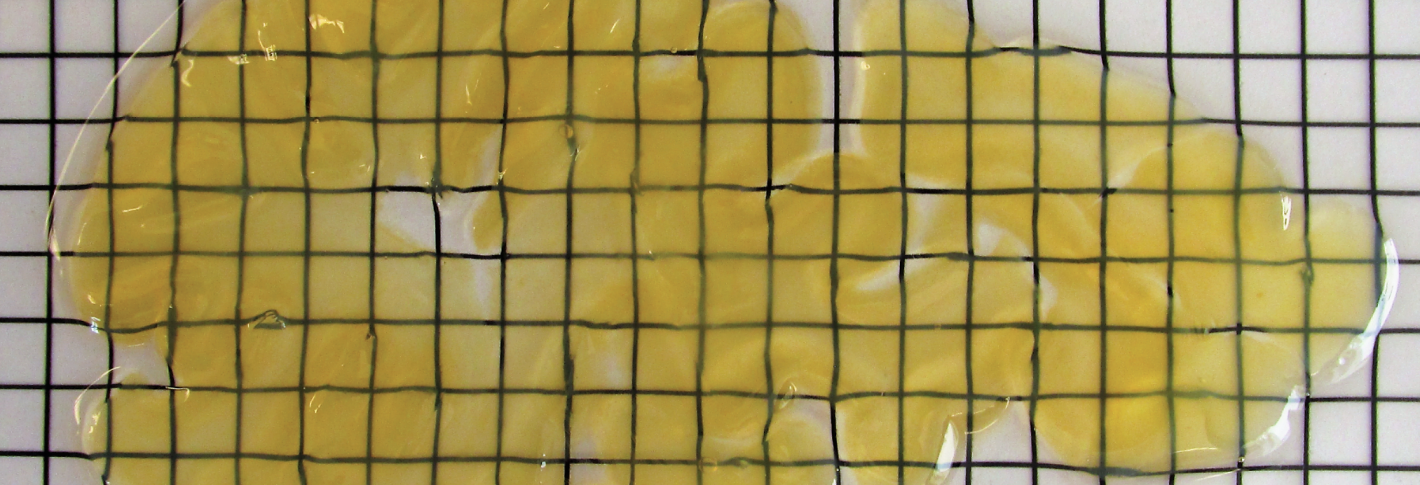
Image above: A 2mm thick slice of human brain, preserved with SHIELD.
Cerebral cartography
The team’s plan for mapping two whole brains will begin at Massachusetts General Hospital (MGH) where collaborator Dr. Matt Frosch, a world-renowned pathologist, the director of the MGH brain bank, and the associate director of HST, will carefully examine brains at autopsy to select two samples that are as “normal” as possible. Dr. Van Wedeen, an associate professor of Radiology at Harvard University and MGH, will then scan the intact brains with MRI to resolve their overall structure and connectivity.
From there the brains will travel to MIT where Chung’s lab will slice them into 2 millimeter-thick slabs (yielding about 60 per brain) and then apply a series of innovative tissue processing methods that Chung has devised. SHIELD allows for a brain to be optically cleared while protecting the structure of every cell and protein and preserving their ability to react with a wide variety of fluorescent labeling agents applied using another chemical process called SWITCH. With the slabs preserved and labeled, the team will be able to scan them using custom-built microscopes with subcellular resolution but also high throughput. After the slabs are scanned, the team will zoom even further in on key areas of interest using MAP, another Chung invention that allows for tissues to be enlarged so that their tiniest structures become resolvable to microscopes.
All these incredibly detailed yet full-scale images will produce an enormous amount of data – Chung estimates about 40 petabytes, (40 million gigabytes). That’s why team members Sebastian Seung at Princeton, Laura Brattain at MIT’s Lincoln Laboratory, and a group of computer scientist in the Chung lab are working together to develop special image processing algorithms.
In all, the project will incorporate many of Chung’s best tissue processing inventions and lead to the invention of even more new technologies. That’s what it takes to map the human brain, which is 2,000 times the volume of a mouse brain.
“It’s something that we have been preparing since I joined MIT,” Chung said. “Now I feel like we are ready to attack this challenge.” The development of the technologies was supported by the JPB Foundation, NCSOFT Cultural Foundation, the Burroughs Wellcome Fund Career Award at the Scientific Interface, the Searle Scholars Program, the Packard Award in Science and Engineering, the McKnight Foundation Technology Award, and the National Institutes of Health.